
- Remember me Forgot password?

Bisulfite sequencing is regarded as the gold standard for detecting the DNA methylome. This analysis pipeline is designed to analyze methylation sites in WGBS data.
This pipeline is developed by the BMAP team.
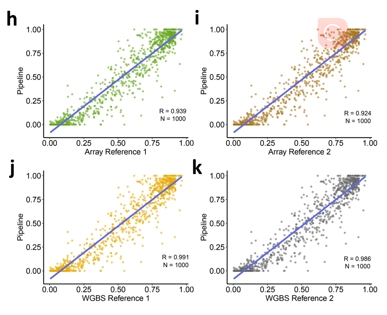
A set of human prostate cancer cell line (LNCa, PrEC) datasets that were detected using both WGBS (Illumina HiSeq 2500) and microarray (Infinium MethylationEPIC BeadChip) was used to assess the accuracy of the WGBS analysis pipeline. These original datasets were downloaded from NCBI-SRA/GEO (SRP089722, GSE86833). We randomly selected 1,000 CpG sites from the over 700,000 CpG sites covered by the original dataset and performed a correlation analysis between the results of the user-customized WGBS analysis pipeline and the two reference pipelines from the original publication.
The correlation coefficient indicates the accuracy of the customized software. Next, we tested the accuracy of the WGBS analysis pipeline in BMAP and obtained an average correlation of 0.960.
The standardized Whole genome bisulfite sequencing (WGBS) analysis pipeline for methylation data was established in BMAP following the guidelines in Krueger[1] and Akalin’s[2]studies.
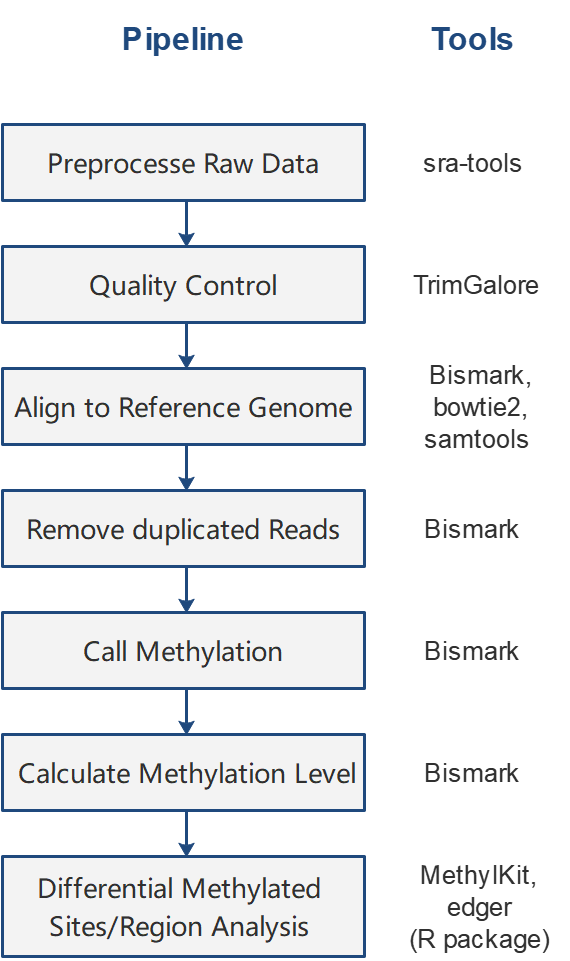
[1] Krueger, F. and Andrews, S.R. (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics, 27, 1571-1572.
[2] Akalin, A., Kormaksson, M., Li, S., Garrett-Bakelman, F.E., Figueroa, M.E., Melnick, A. and Mason, C.E. (2012) methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol, 13, R87.
 Address: 800 Dong Chuan RD. Minhang District, Shanghai, China SJTU-Yale Joint Center for Biostatistics, SJTU
Address: 800 Dong Chuan RD. Minhang District, Shanghai, China SJTU-Yale Joint Center for Biostatistics, SJTU
Copyright © 2021 沪交ICP备20190249. All Rights Reserved