
- Remember me Forgot password?

RNA-protein binding sites prediction suite based on deep learning
This webserver is provided by the author of the Hong-Bin Shen‘s group[1].
RNA-binding proteins (RBPs) are involved in many biological processes, their binding sites on RNAs can give insights into mechanisms behind diseases involving RBPs. Thus, how to identify the RBP binding sites on RNAs is very crucial for follow-up analysis, like the impact of mutations on binding sites. With high-throughput sequencing developing, there is an explosion in the amount of experimentally verified RBP binding sites, e.g. eCLIP in ENCODE. However,CLIP-seq to detect RBP binding sites relies on gene expression which can be highly variable between experiments, and cannot provide a complete picture of the RBP binding landscape. Computational methods are in urgent needed to predict missing binding sites for individual RBPs.
RBPsuite mainly contain two deep learning-based approaches, iDeepS and iDeepC for linear RNAs and circular RNAs. iDeepS is deveoped for predicting RBP binding sites on linear RNAs. iDeepC is developed for predicting RBP binding sites on circular RNAs.
This benchmark dataset consists of 353 RBPs and their binding sites (RNAs) are derived from POSTAR3 database. Click here (722M) to download the benchmark dataset for RBP binding linear RNAs of RBPSuite.
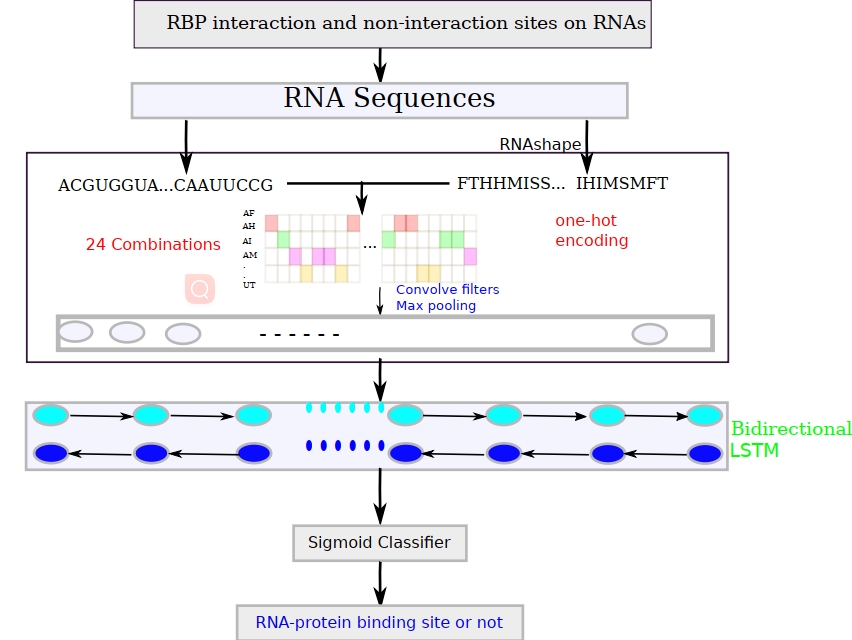
Figure. The flowchart of updated iDeepS for predicting RBP binding sites on linear RNAs
[1] Xiaoyong Pan, Yi Fang, XianFeng Li, Yang Yang, and Hong-Bin Shen. RBPsuite: RNA-protein binding sites prediction suite based on deep learning. BMC Genomics, 2020, 21:884.
 Address: 800 Dong Chuan RD. Minhang District, Shanghai, China SJTU-Yale Joint Center for Biostatistics, SJTU
Address: 800 Dong Chuan RD. Minhang District, Shanghai, China SJTU-Yale Joint Center for Biostatistics, SJTU
Copyright © 2021 沪交ICP备20190249. All Rights Reserved