
- Remember me Forgot password?

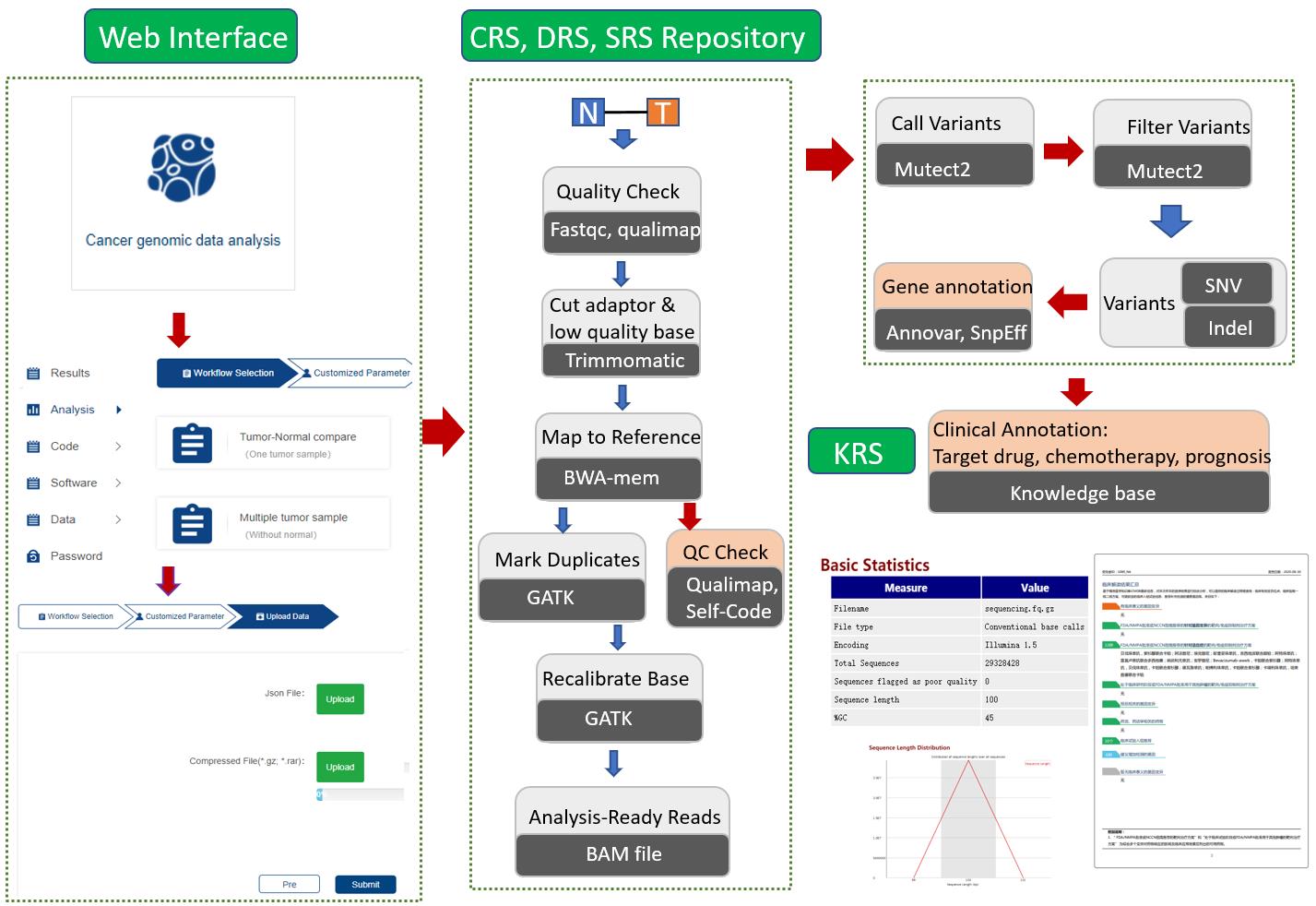
This cancer genomic data analysis platform was developed based on the guidelines of Mutect2. Afterwards, the variants were annotated following the HGVS guidelines, and the clinical annotations were inferred from the clinical knowledge graph iCMDB.
This pipeline is released by the BMAP team.
This pipeline was validated using a standard sample (GW-OYC001), which consists of 19 target drug-related genes and 21 mutations, with mutation frequencies ranging from 5% to 60%. The pipeline successfully detected all variant sites, and all variants in the standard sample were cross-validated using both NGS and ddPCR.
NGS based somatic variant calling and clinical annotation.
This application processes NGS sequencing data to identify somatic variants and generates an assistant report, which includes treatment recommendations and relevant clinical trial options.
 Address: 800 Dong Chuan RD. Minhang District, Shanghai, China SJTU-Yale Joint Center for Biostatistics, SJTU
Address: 800 Dong Chuan RD. Minhang District, Shanghai, China SJTU-Yale Joint Center for Biostatistics, SJTU
Copyright © 2021 沪交ICP备20190249. All Rights Reserved