
- Remember me Forgot password?

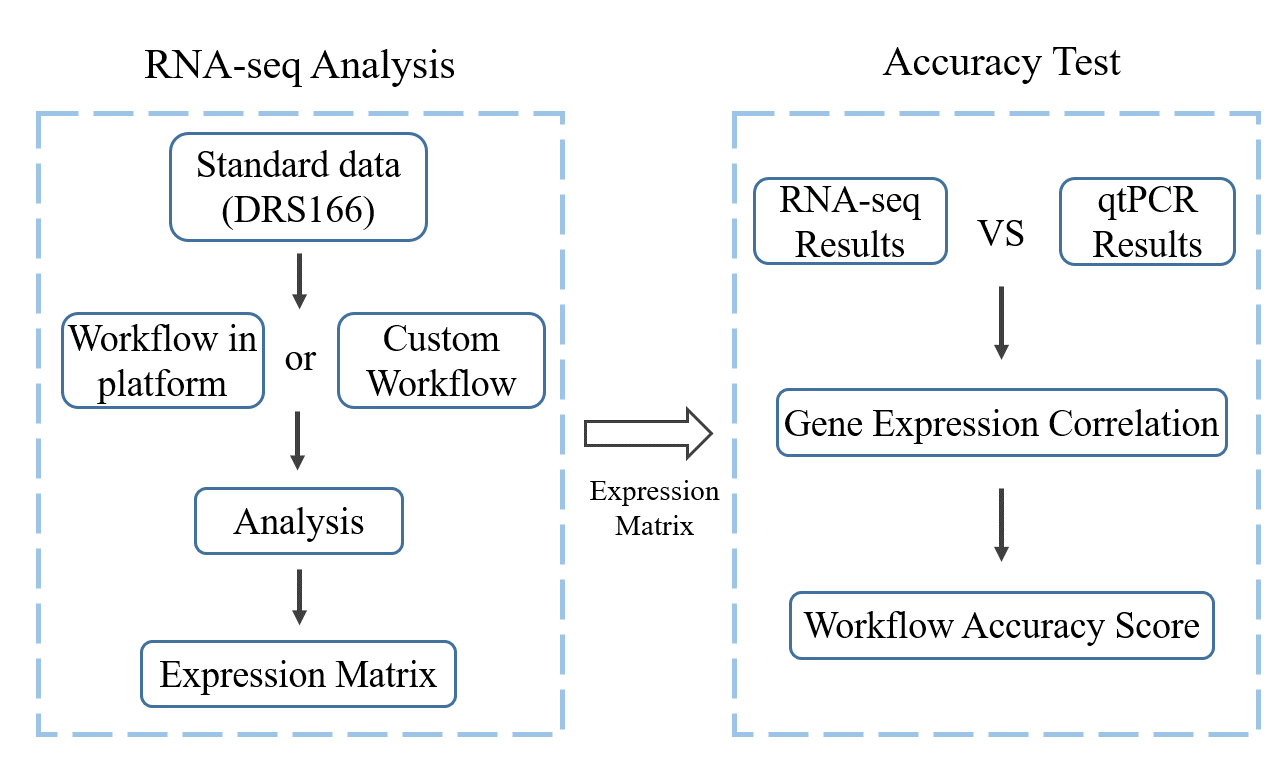
RNA-seq is an important technology for cell expression research. Hundreds of analysis tools have been developed in recent years. Each tool has its applicable conditions, solide part and weak links, which leads to a wide variety of existing RNA-seq processes and large differences in results. There has been some research work that has evaluated the accuracy of the latest available tools. However, there is no comprehensive assessment of the overall accuracy of the RNA-seq process. Therefore, we collected the transcriptome sequencing data of 8 samples and the corresponding qtPCR data of 1000 genes based on the data in the literature, providing a feasible method for testing the accuracy of the transcriptome analysis process. Currently, it supports the correlation and accuracy detection of the platform's existing and user-provided transcriptome analysis workflows. This method quantifies the detection results and supports horizontal comparison, which makes the accuracy and stability of each process more intuitive to provide scientific researchers.
First, use the RNA-seq process to be detected to analyze the DRS575016784630132736 data, and then the gene expression data file is input the detection process to obtain the accuracy of the process.
 Address: 800 Dong Chuan RD. Minhang District, Shanghai, China SJTU-Yale Joint Center for Biostatistics, SJTU
Address: 800 Dong Chuan RD. Minhang District, Shanghai, China SJTU-Yale Joint Center for Biostatistics, SJTU
Copyright © 2021 沪交ICP备20190249. All Rights Reserved